Research Overview.
As a deep-sea biologist, I use DNA sequencing and genomic technologies to study the biodiversity, ecology, and evolution of microbial species that inhabit seafloor sediments (mainly, nematode worms that live in ocean mud). My research lab website can be found here, which includes a full list of my peer-reviewed scientific publications.
Here are a few of the specific topics which I’m most excited about lately:
Ecology & Evolution of Deep-sea nematodes
What can nematode worms teach us about the origin and persistence of animal life in the deep ocean?
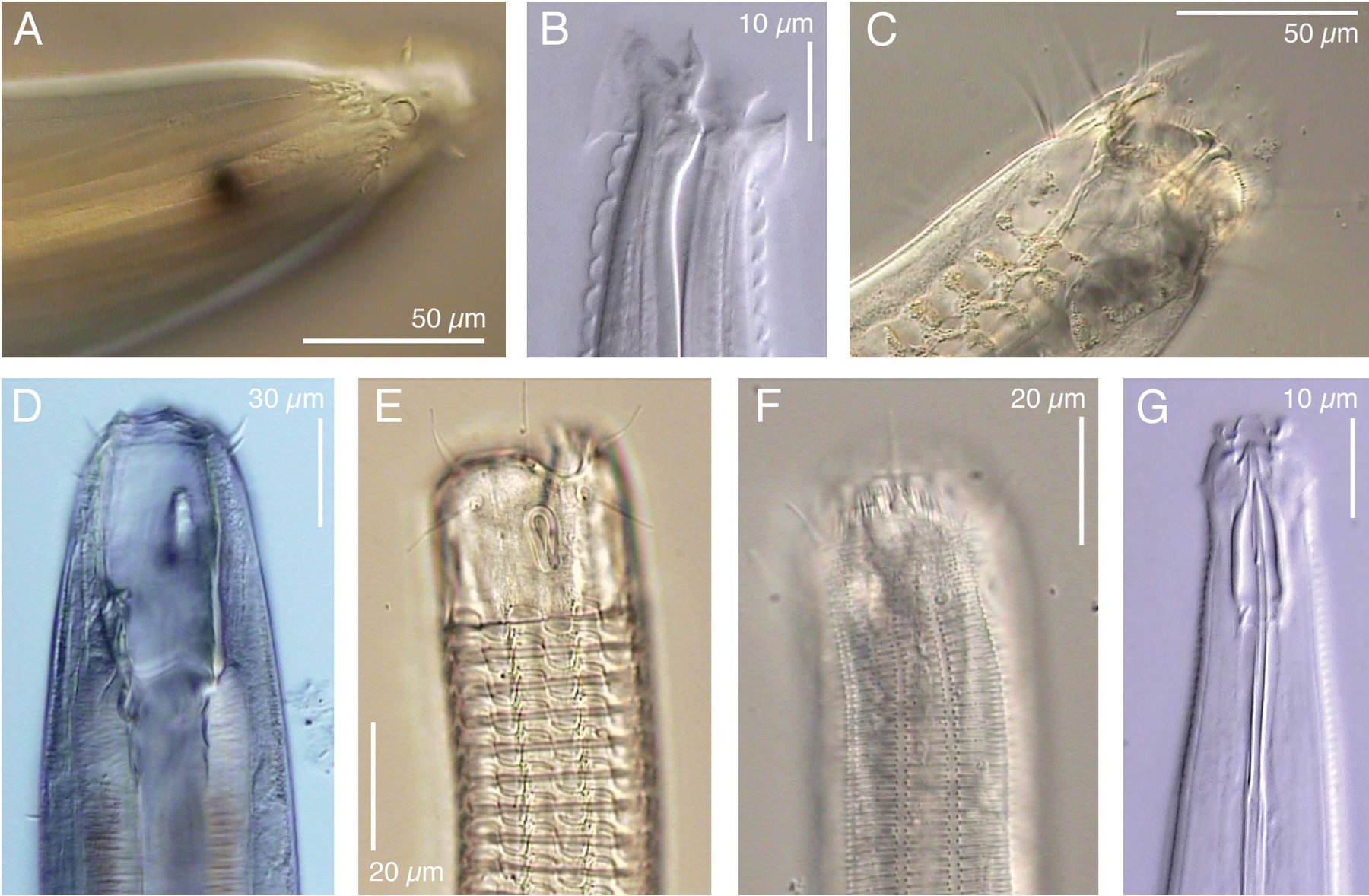
Nematode worms (roundworms) are microscopic animals that inhabit nearly every type of soil, sand, and mud found around the world - from high-altitude volcanic slopes down to the deepest ocean trenches. Free-living nematode species are generally less than 1 millimeter in length, and their biodiversity is purported to rival that of insects (millions of species). This important group of worms is notoriously understudied, and to date, taxonomists have described less than 5,000 total species from marine environments. Most scientific research on nematodes has historically focused on terrestrial species (agricultural pests such as root-knot and potato-cyst nematodes), parasites of human and livestock (dog heartworm!), and the model lab species Caenorhabditis elegans.
My research aims to rapidly describe new marine nematode species from around the world — focusing especially on deep-sea ecosystems — using a mixture of DNA sequencing, evolutionary trees (phylogenetics), and formal taxonomic species descriptions (via high-resolution microscopy and imaging). Deep-sea habitats cover 91% of the earth’s surface (sediments > 200m depth), but our current understanding of deep-sea nematode species is based on a cumulative sampling area less than half the size of a tennis court (sediment cores totaling 60-70 sq. meters of seabed). I am especially interested in the concept of marine nematodes as "evolutionary commuters" that can colonize new habitats on short timescales due to their simplistic body plan and cellular physiology.
(Photo from De Ley 2005, WormBook)
Invertebrate microbiomes
What governs the community assembly of nematode microbiomes - microbial metabolism, natural selection, or chance?
Over the last decade, an explosion of scientific studies have revealed the critical biological importance of animal-associated microbiome taxa (species of bacteria, archaea, protists and fungi). However, our knowledge of invertebrate microbiomes is still relatively scarce compared to studies of vertebrates. Unlike more complex animal groups such as mammals, invertebrate species do not appear to exhibit "phylosymbiosis" where microbiome taxa mirror the evolutionary tree of the animal host. Yet, invertebrate species still seem to harbor microbiome communities that are definitively distinct from the mixture of microbes present in their environmental surroundings.
My lab is helping to drive forward the emerging research area of nematode microbiome studies. Recent evidence suggests that closely related nematodes species may maintain separate microbiome niches to avoid competition, and other studies have further detected seasonal turnover in dominant bacterial symbionts of marine nematodes. I am broadly working to characterize the species composition and functional gene content of diverse nematode microbiomes, in order to understand how host-associated microbial communities may help to drive the evolution and ecology of diverse marine nematode species.
(Photo from Bellec et al. 2019, Scientific Reports)
Antarctic MaRine Biodiversity
Are Antarctic marine sediments “hotspots” of biodiversity for microscopic animal species?
The Antarctic continental shelf is the only known marine ecosystem where deep-sea fauna emerge into shallow waters. This is most likely a result of the extremely dense, cold, and salty ocean waters in the Southern Ocean which allow easy migration up from the depths (versus other parts of the world where warmer temperatures in the shallows act as a formidable barrier). Antarctic marine invertebrates appear to have wider depth ranges compared to their close relatives in other ocean basins, and the Southern Ocean is home to high numbers of endemic species not found anywhere else in the world (most likely due to the remote geographic location and relative isolation of this water mass as a result of the ferocious circumpolar current that encircles the entire Antarctic continent). Historical cycles of glacier formation and ice shelf melting in Antarctica have also contributed to unique evolutionary and ecological conditions not found in any other marine ecosystem in the world.
I am interested in applying modern genomic tools to study microscopic animals (and their microbiomes!) in Antarctic marine ecosystems. We do not have robust DNA sequence databases for most small invertebrate species in Antarctica, with very little molecular data in existence for species-rich groups such as nematode worms. This lack of baseline information about Antarctic ecosystems is a critical issue given the rapid warming of the Southern Ocean and the imminent threat of climate change in this unique marine ecosystem.
(Photo via Unsplash)